Yet Another Bayesian Networks Library
Over the past month, apart from working on Cim and going out I have worked a little on a project on which I had been fantasying a lot for months : a bayesian network library.</p>
There are already quite a few open-source bayesian network libraries available out there. However, those written in Java are only released under the GPL (see BNJ, JavaBayes and RISO). As I’m usually looking for LGPL libraries when it comes to integrating foreign code to mine (so as to be able to release my code under both a proprietary license and the GPL), I decided to write my own library. This vague intent dates back to last december, and to prepare this huge project I bought a very nice book about bayesian networks when I travelled to France to get my engineering degree. Since then I’ve been reading that book on and off, until I eventually decided to implement a few basic components needed by bayesian networks computations.</p>
I started with a simple yet rather efficient general-purpose graph library. I chose to represent a graph using a matrix, or more precisely a sparse matrix. Indeed, it is very easy to do advanced computations on a graph once you get its adjacency matrix (of size N x N if the graph has N nodes). This launched a subproject to build a small yet functional and efficient sparse matrix / sparse vector library, with maximal optimizations for sparse matrices multiplications and additions. To keep the memory footprint of this library as small as possible, I banned the use of java.util.HashMap and java.util.TreeMap and wrote specialized int -> Object and int -> int sorted array maps.</p>
The sparse matrix library allowed me to implement some graph algorithms with a cheap computational cost, once the full connectivity matrix is computed. For instance, asking to a graph if there is a path between two nodes is made in constant time.</p>
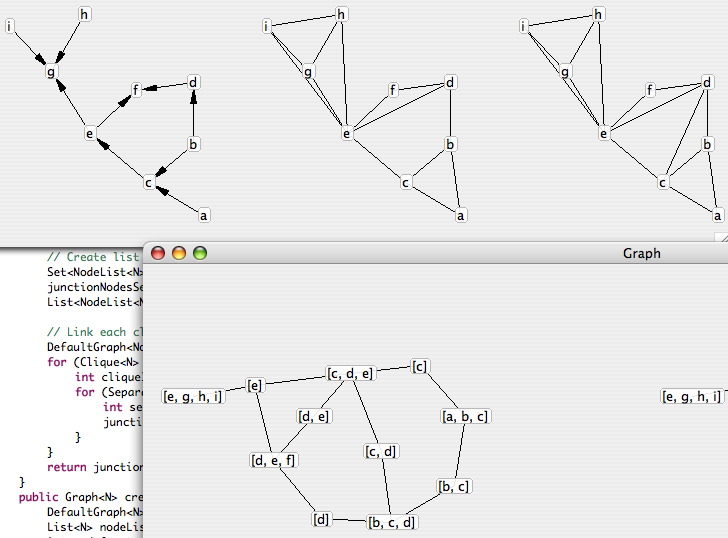
Here are a few features of my graph library in its current state :</p>
- get all the children / descendents, parents / ancestors of a node
-
parse a human-readable string that describes a graph, such as “a->b->c< -d e<->a” (oriented) or “a-b-c-d-a b-d” (non oriented) </li>
- get all the paths between two nodes
- get all the cliques of a graph
- get the properties of the graph : isAcyclic, isTree
- moralize an oriented graph
- triangulate a non oriented graph
- get the junction graph of a non oriented graph
- test active chains and ask if two sets of nodes are D-separated by a third one in an oriented graph
I also made a small graph display component that tries to find the best position for all nodes, possibly taking into account several graphs that have the same nodes list (useful to display graphs that were obtained from each other with a consistent node layout). This component uses a cost function that contains attractive potentials for linked nodes, repulsive potentials at small distance to avoid a collapse of the graph and angular repulsion to get nice graphs. It still misses penalties for edges intersections but that should come later. It uses a simple gradient descent + simulated reheat and is very far from being efficient, but it already produces acceptable results in most cases (my goal was just to check my algorithms are applied correctly without having to decipher a text dump)..
To finish with graphs now I only need to implement the minimalization of a junction graph (removal of redundant edges in cycles where the intersection of separators is non null).</p>
Then the real stuff can begin : I already started a small discrete random-variables computation library, with support for conditional likeliness tables, functions with arbitrary number of variables, arbitrary function sums and products with constant optimizations and inner functions’s arguments gathering, marginalization and instantiation of sets of variables + standard evaluation. This is meant to handle the probabilistic functions that will be generated to represent the cliques’s potentials as computed from the minimal junction graph of a triangulated moralized graph of an arbitrary oriented graph.</p>